Documentation
Background
Post-translational modifications (PTMs), such as phosphorylation, glycosylation, methylation, and ubiquitination, are key regulators of protein function and structure. One fundamental step towards understanding the role of genetic variants in disrupting protein function is the ability to map and inspect their positions and enrichments, relative to PTM sites. Missense3D-PTMdb is a freely available platform that provides comprehensive identification of missense variants at or near PTM sites. The platform provides users with a user-friendly interface to query a single residue position within a canonical protein, returning detailed variant and PTM annotations.
How to use Missense3D-PTM
On the Home page, users can submit an input form to query a position within their protein of interest. Here, users must input:
- (1) UniProt ID OR Gene Name: The primary accession UniProt ID or recommended gene name for the protein of interest
- (2) Residue Position: The position within the fasta sequence of the canonical protein of interest from UniProt
- (3) Wild Type Residue: The wild type residue at the position inputted for the canonical protein of interest from UniProt
Understanding your results
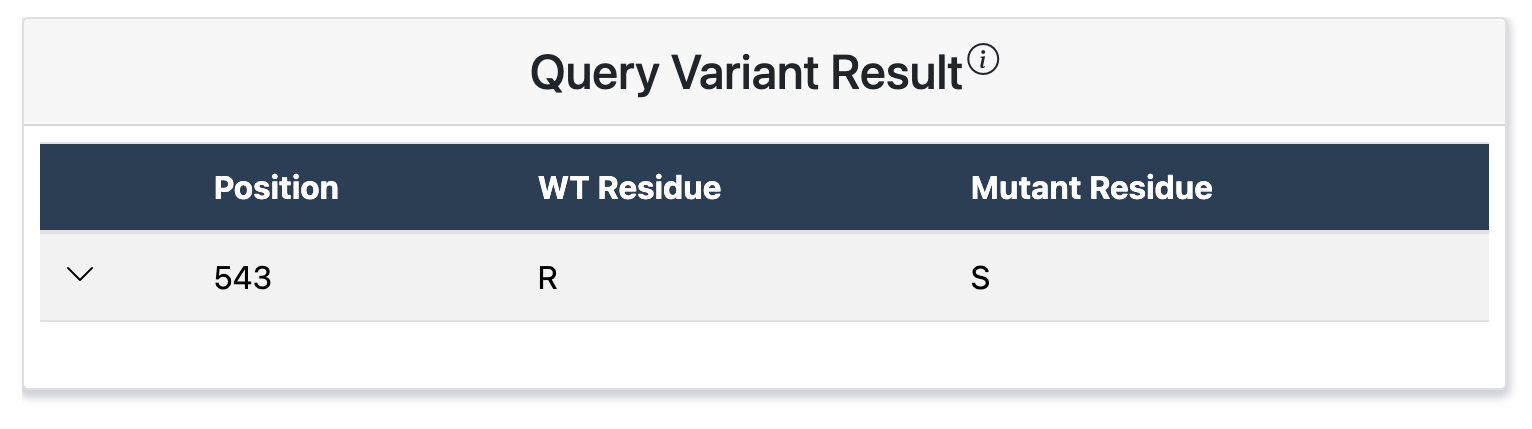
Variant Results
A single query returns, if any, existing missense variants identified at the specified position on the canonical protein of interest. Variants are organized by mutant allele, such that a single row contains the queried position and wild type (WT) allele for a single variant alongside the respective mutant allele.
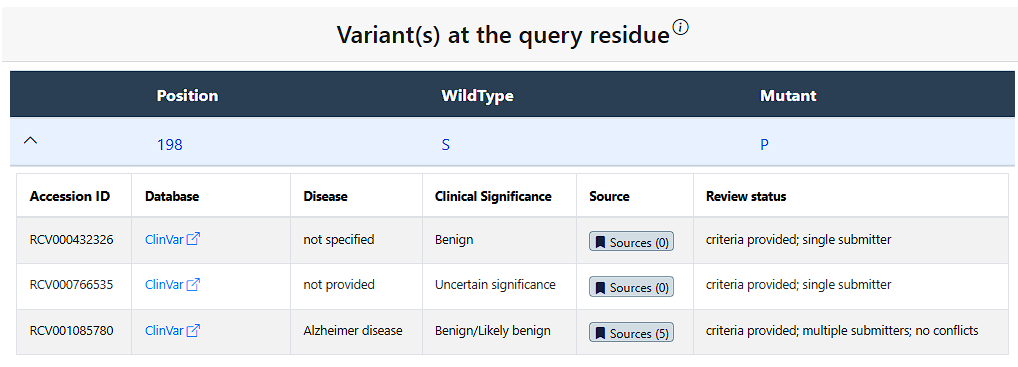
Clicking the drop down arrow reveals all documented missense variant records for each mutant allele occuring at the queried position. A single mutant allele can have numerous records associated with it, which are respectively displayed in individual rows. For each record, the table provides information on:
- (1) Accession ID - The accession code used for this variant in these Databases
- (2) Database - The corresponding database(s) the variant record is found in, note the text can be clicked to see these database's pages for this variant
- (3) Disease - Any associated disease reported for the variant record
- (4) Clinical Significance - Any predicted or determined clinical pathogenicity associated with the variant
- (5) Source - PubMed source link(s)
- (6) Review status - The review status of the variant record where provided
PTM Results
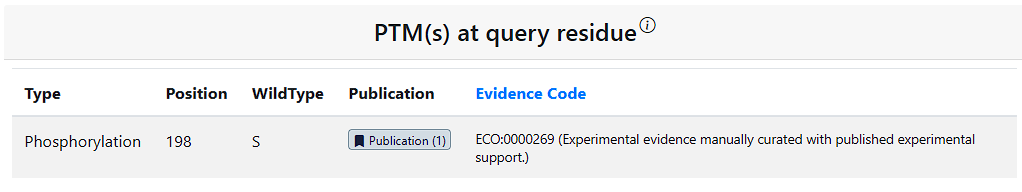
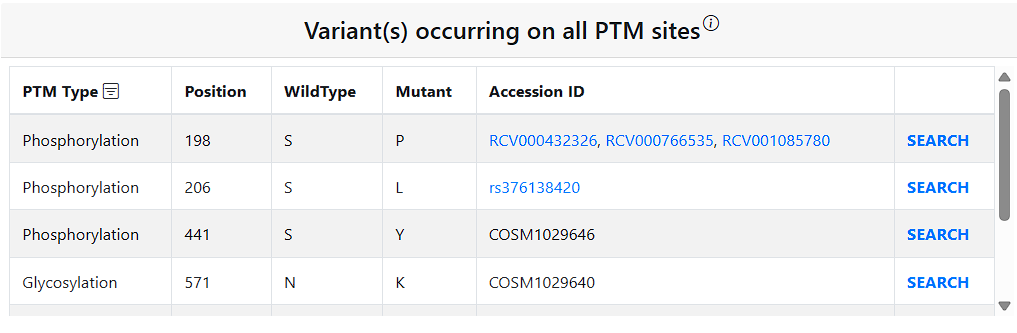
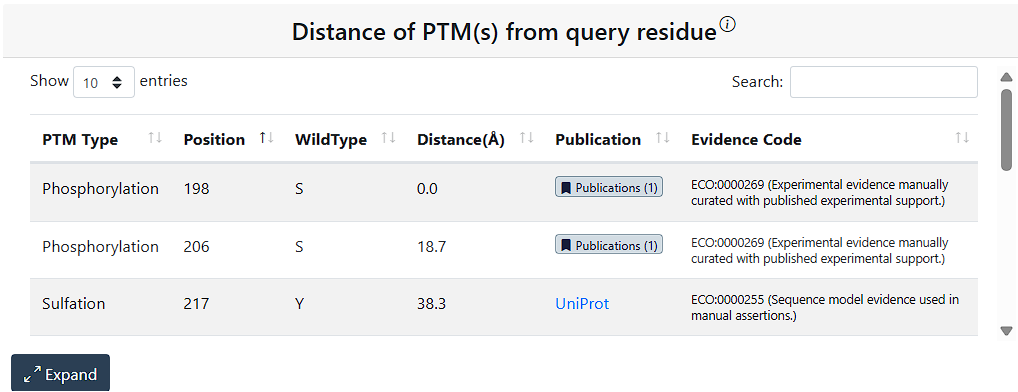
To facilitate users’ research on PTM–variant relationship from a broader perspective, the Results page includes additional relevant information on PTMs and variants. The PTM(s) at query residue table shows information for any PTMs on the query residue. The Variant(s) occurring on all PTM sites table show information on all PTM sites in the protein that have a missense variant. The Distance of PTM(s) from query residue shows all PTMs in the query protein and their distance from the query residue in angstroms.
Protein Structure Information
To aid interpretation of PTMs and variants in 3D structure a visualisation of the AlphafoldDB model of the protein is displayed using JSmol a feature complete 3D viewer. This supports all standard JSmol features and scripting more information can be accessed here.
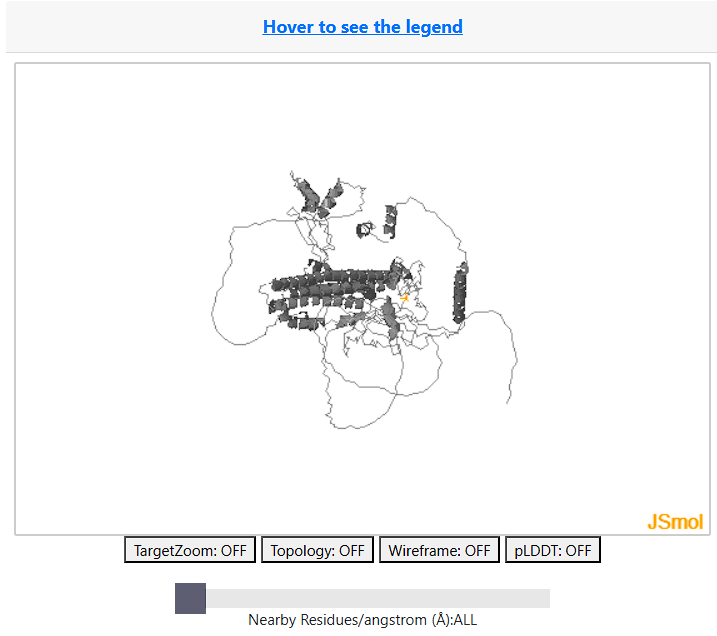
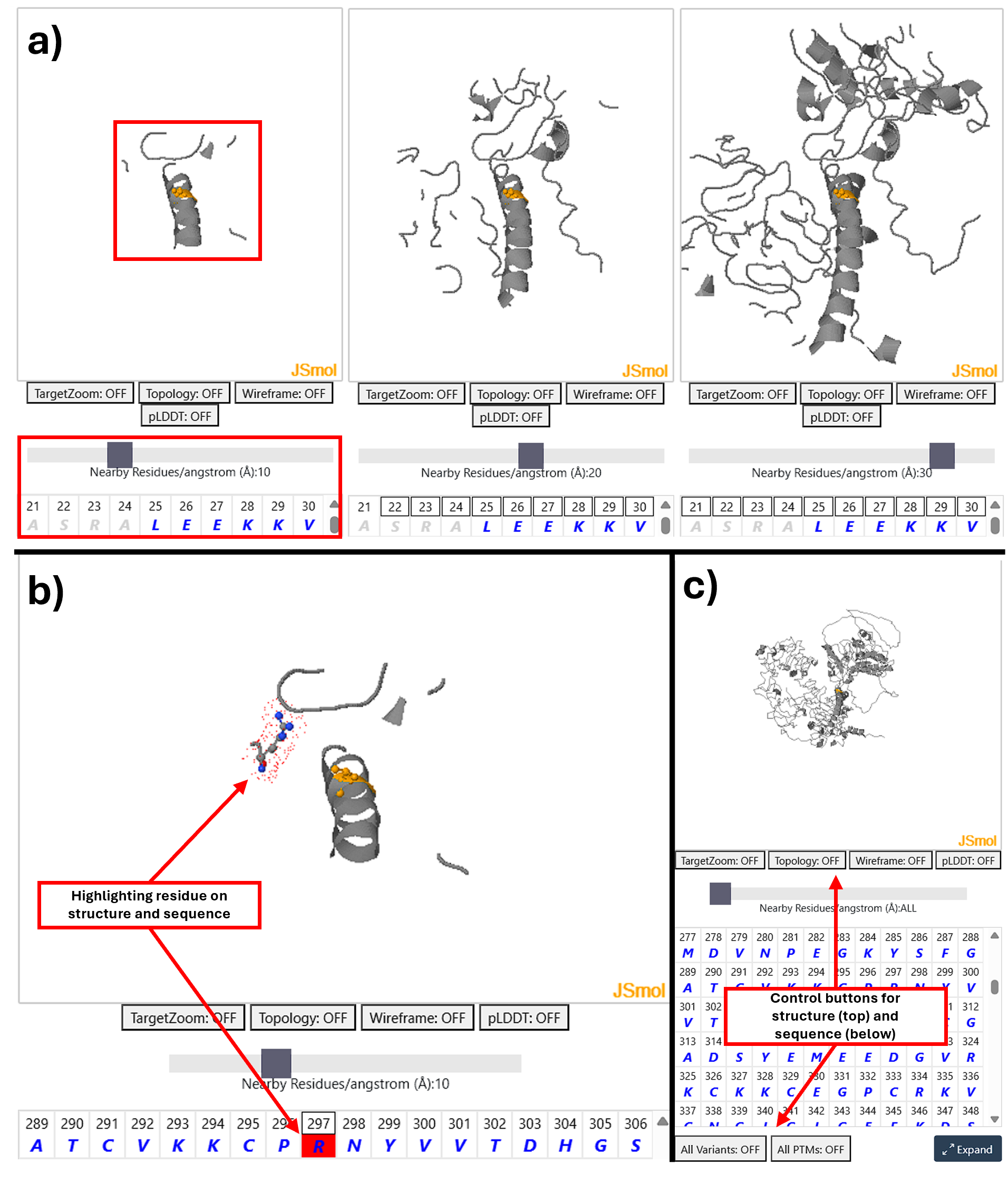
Note: The JSMol Viewer will not dispaly proteins longer than 2,700 amino acids.
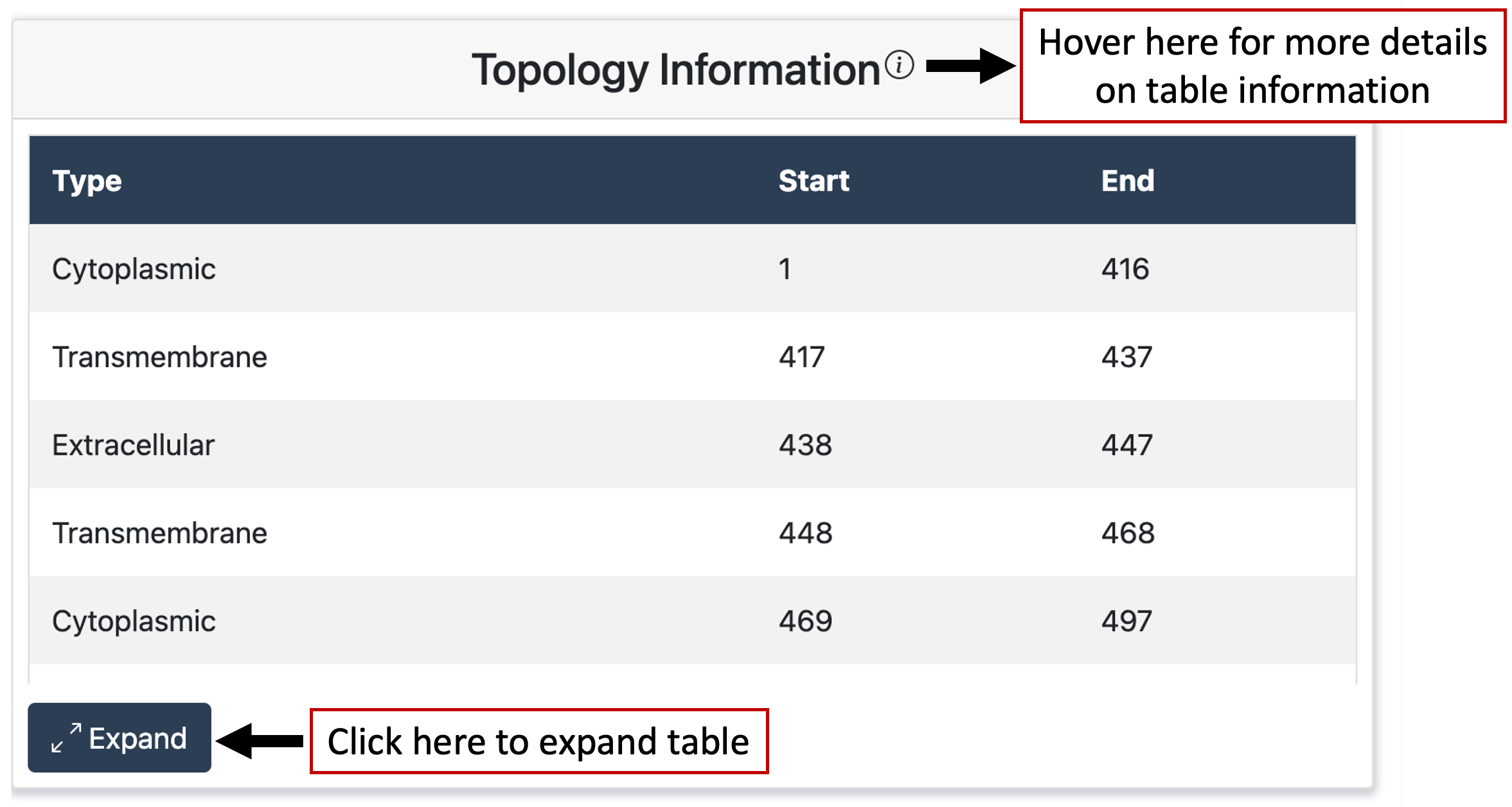
Topology Results
Results also display topology information for the queried protein. The table displays the positions of the topological domains as well as the domain type, if it exists, for the protein.