
Check if your structure contains a sequence that is up-to-date with the UniProt entry.
UniProt entries describing protein sequence are regularly updated as researchers collect new sequencing evidence.
Protein structure files (modelled or experimental) often lag behind these sequence updates. Sequences within structure files might not fully represent assigned UniProt entries and the residue numbers might not match the UniProt numbering.
Mapping UniProt annotations (i.e. variants, domains, binding sites) to outdated structure files may result in errors and hinder downstream structural analysis.
3DSeq-Check checks your structure file against the given UniProt entry. 3DSeq-Check gives maps your structure to the current sequence of the UniProt entry and gives an overview of the mapping.
1. Input description
UniProt ID is the first required input to StructureCheck-UP. The UniProt ID should correspond to the entry that your structure describes, it is used to fetch the current up-to-date sequence that is used as a reference sequence in the StructureCheck-UP.
Structure Source is the second required input to StructureCheck-UP. The structure source should correspond to the structure you want to check-UP. There are currently two supported ways to supply a structure:
- AlphaFoldDB - by choosing AlphaFoldDB as source, structure file corresponding to the suplied UniProt ID will be fetched and used for comparison.
- Custom PDB File - by choosing a custom file as source, the file you upload will be used as input. In addition to the PDB file you also need to provide the chain id for the chain you want to check-UP, this should be a chain that corresponds to the supplied UniProt ID.
2. Extracting the sequence
To fetch the up-to-date reference sequence, we use the UniProt REST API for the entry with the input UniProt ID. The query sequence is extracted from the structure source, by extracting the residues that appear in the `ATOM` section of the structure file (and with the matching chain id, in the case of custom file input).
3. Sequence alignment
The pairwise sequence alignment of the reference and query sequences is performed using the EMBOSS implementation of the Needleman-Wunsch algorithm with the default parameters.
3. Output visualization
Resulting alignment of the query sequence (orginating from the input structure source) to the refernce sequence (the up-to-date UniProt sequence) is presented in the results page.
- A summary table is displayed on top. The summary table shows:
- length of aligned sequences
- sequence identity between the query sequence (structure) and the reference sequence (UniProt)
- number of gaps (insertions/deletions to the reference UniProt sequence as compared to the query sequence) in the alignment
- number of variations (different amino acids between the reference UniProt sequence and the query sequence) in the alignment
-
Match assessment message is displayed bellow the summary table and contains three possible assessments:
- Your structure matches the UniProt sequence, safe to use! . is displayed in case that the sequence identity 100% and the UniProt sequence fully matches the sequence in the structure. This corresponds to a perfect alignment and the recommendation is to freely use the UniProt annotations for the supplied UniProt ID on this structure.
- Some differences between the UniProt sequence and structure - use carefully. is displayed in case that the sequence identity is above the 50% theshold but less than 100% . This corresponds to an alignment of varying quality and the recommendarion is to use the UniProt annotations carefully for the supplied UniProt ID on this structure.
- Warning: Very poor match between the structure and UniProt sequence! is displayed in case that the sequence identity is below the 50% theshold. This corresponds to a very poor alignment and the recommendation not to use the UniProt annotations for the supplied UniProt ID on this structure.
- Alignment is presented in an interactive dashboard using the Nightingale web components and the insertions/deletions/variations are displayed in separate tracks on top of the alignment.
- Fasta file contents are available for download.
Resulting matching of the residue indices from the protein structure to the indices in the UniProt sequence is presented in the results page.
- A summary table is displayed on top. The summary table shows:
- number of residues in the structure
- number of residues in the structure for which the indices match the UniProt sequence indices
- number of residues in the structure for which the indices do not match the UniProt sequence indices
- number of residues that are missing from the structure and are present in the UniProt sequence
-
Match assessment message is displayed bellow the summary table and contains three possible assessments:
- Your structure residue indexes match the UniProt sequence, safe to map features! is displayed in case that all the residue indexes of the structure match all the indexes of the UniProt sequence. This corresponds to a perfect index match and the recommendation is to freely use the UniProt annotations for those residues that are not missing in the structure.
- Some differences between the UniProt sequence indices and structure indices - map features carefully. is displayed in case that are some mismatch indexes, but that more than 50% of the structure residues match residue indexing of the UniProt sequence. The recommendarion is to use the UniProt annotations carefully for the supplied UniProt ID on this structure.
- Warning: Very low match of residue indices. Be careful when mapping uniprot features. is displayed in case that less than 50% of structure residue indexes match UniProt sequence indexing. This corresponds to a very poor matching and the recommendation not to use the UniProt annotations for the supplied UniProt ID on this structure.
- Alignment is presented in an interactive dashboard using the Nightingale web components and the match/mismatch labels are displayed in separate tracks on top of the alignment.
-
Detailed residue table shows the following information for each residue in the alignment result:
- alignment index - index in the alignment (starting from 1)
- uniprot residue - AA on that position of the UniProt sequence
- uniprot index - index in the uniprot sequence (starting from 1 of the first residue in the UniProt sequence)
- structure residue - AA on that position in the structure file
- structure index - index in the structure file (extracted from the ATOM lines of the structure file)
- label - match/mismatch/missing
- Renumbered structure file for the selected chain is available for download in PDB format.
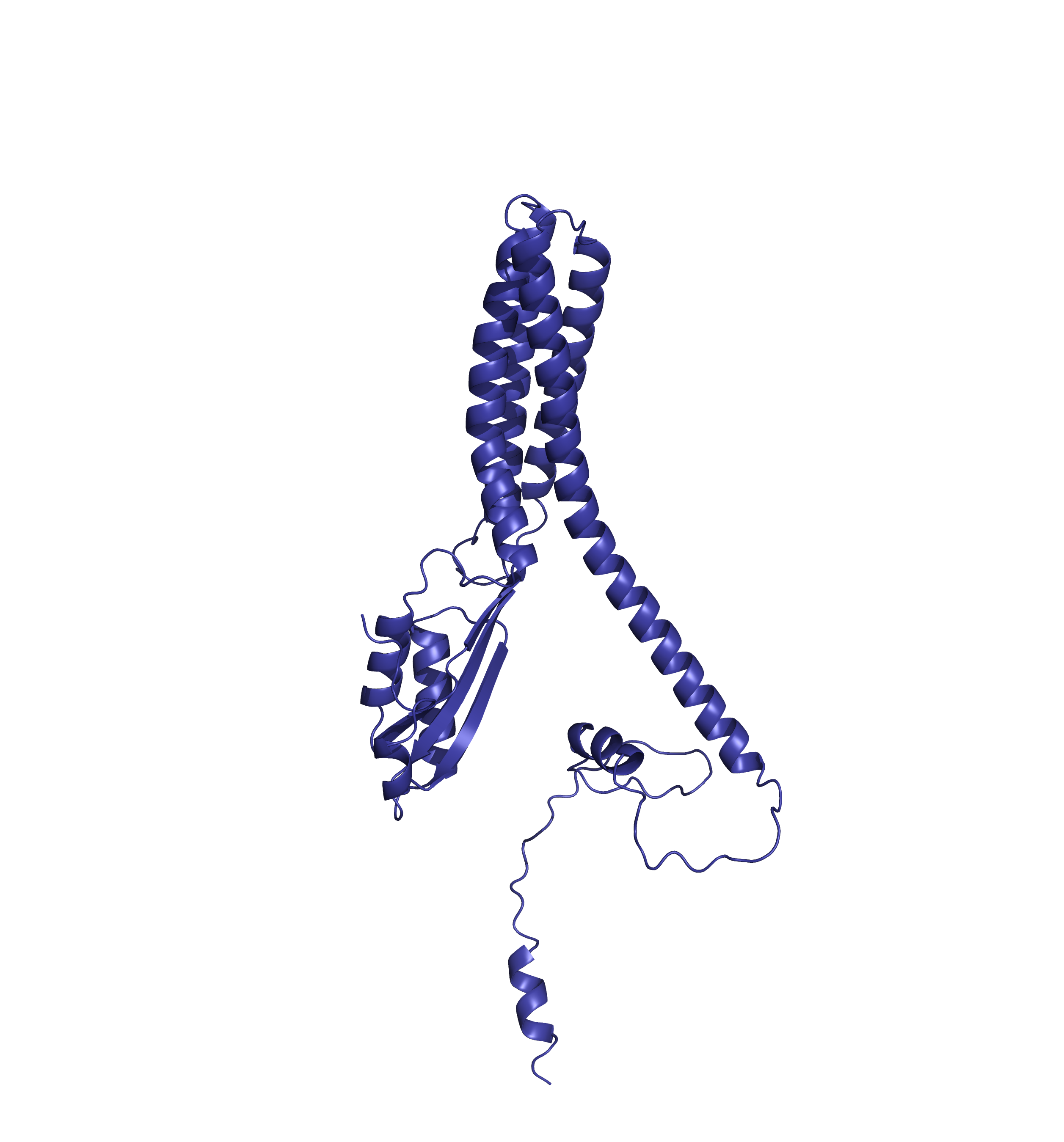
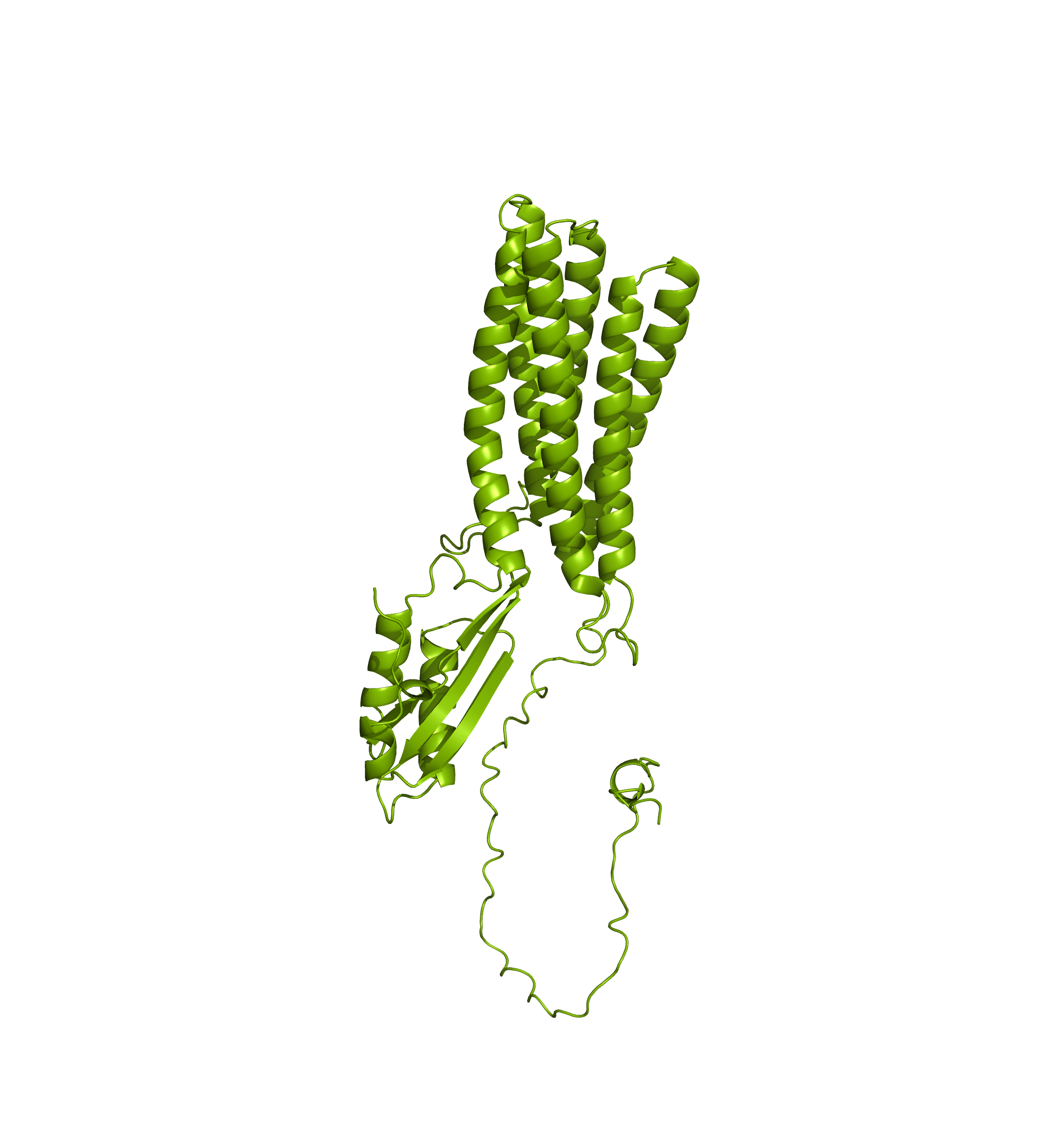
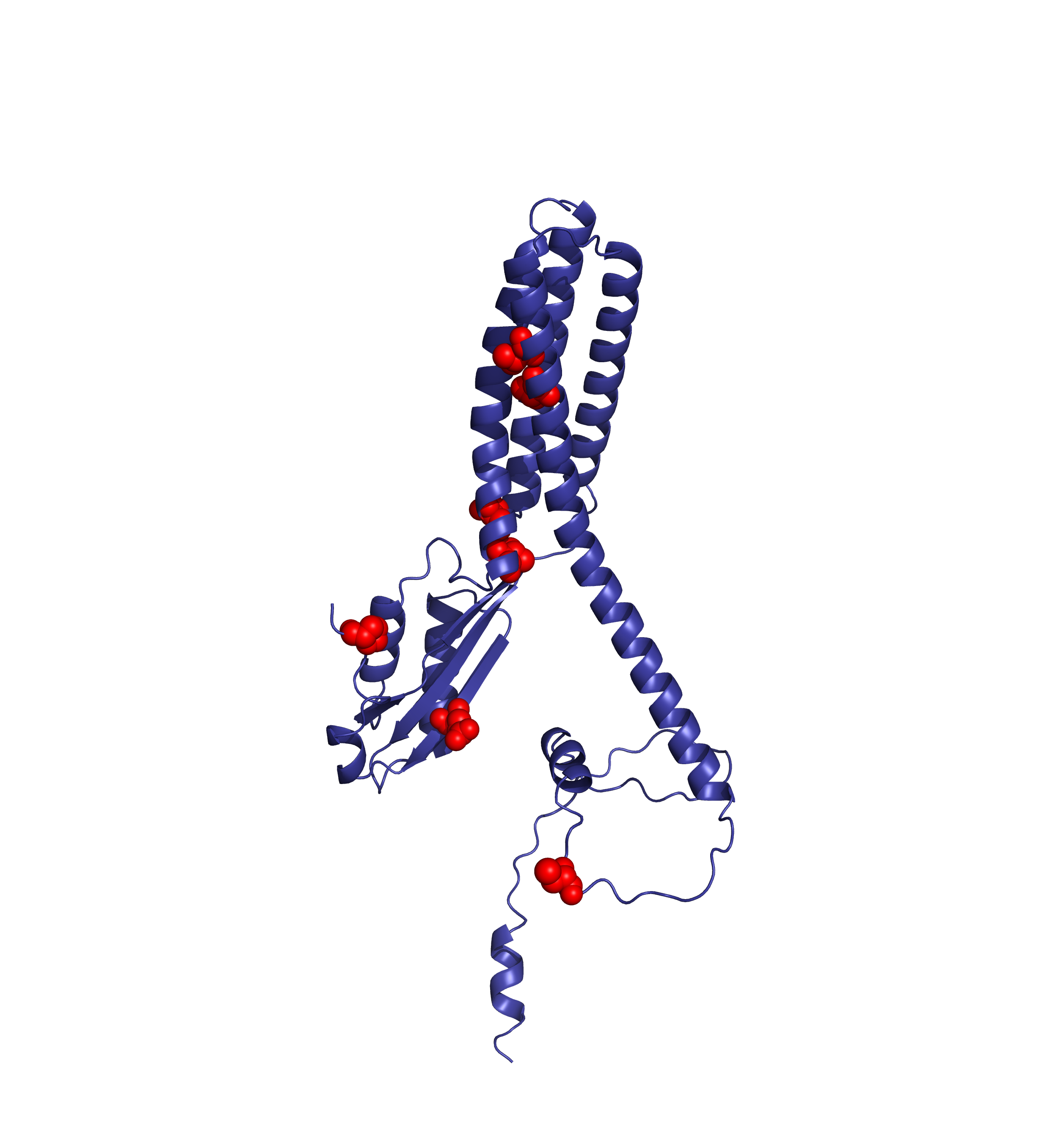
AlphaFoldDB model of proton-coupled zinc antiporter (UniProt ID: Q9BRI3)
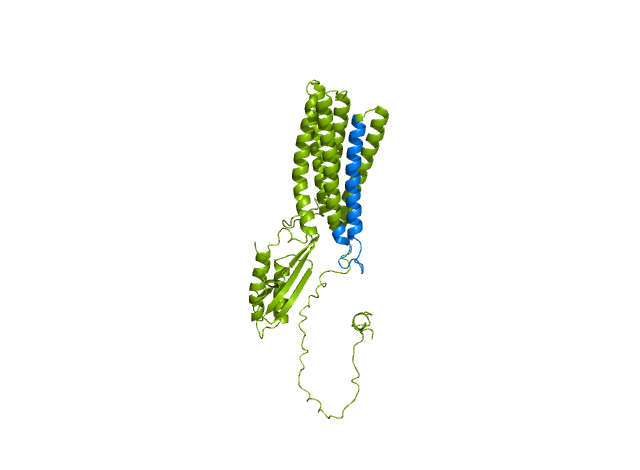
AlphaFoldDB currently (December 2024) stores a model of the UniProt entry Q9BRI3 that reflects version 1 of the Q9BRI3 sequence (changed in October 2022). Since then, an update to the UniProt entry led to a new version of the sequence. Our alignment reveals that version 2 of Q9BRI3 sequence contains ~50 new residues 'inserted' around position 100 of the sequence that was modelled for AlphaFoldDB.
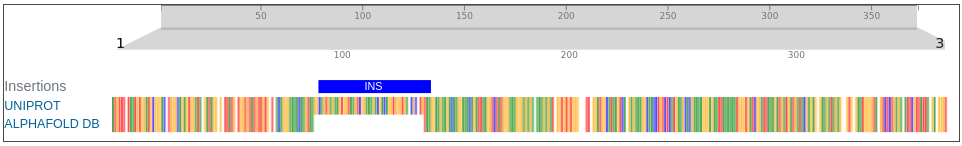
Comparing the structure deposited in AlphaFoldDB (purple, left) and the model of the up-to-date sequence (green, center and right) we see notable differences in the fold. The inserted sequence chunk is highlighted in blue in the figure on the right.
Forthermore, mapping annotations such as Zn2+ binding sites to the outdated models (highlighted below with red spheres) might result in misleading structural views.
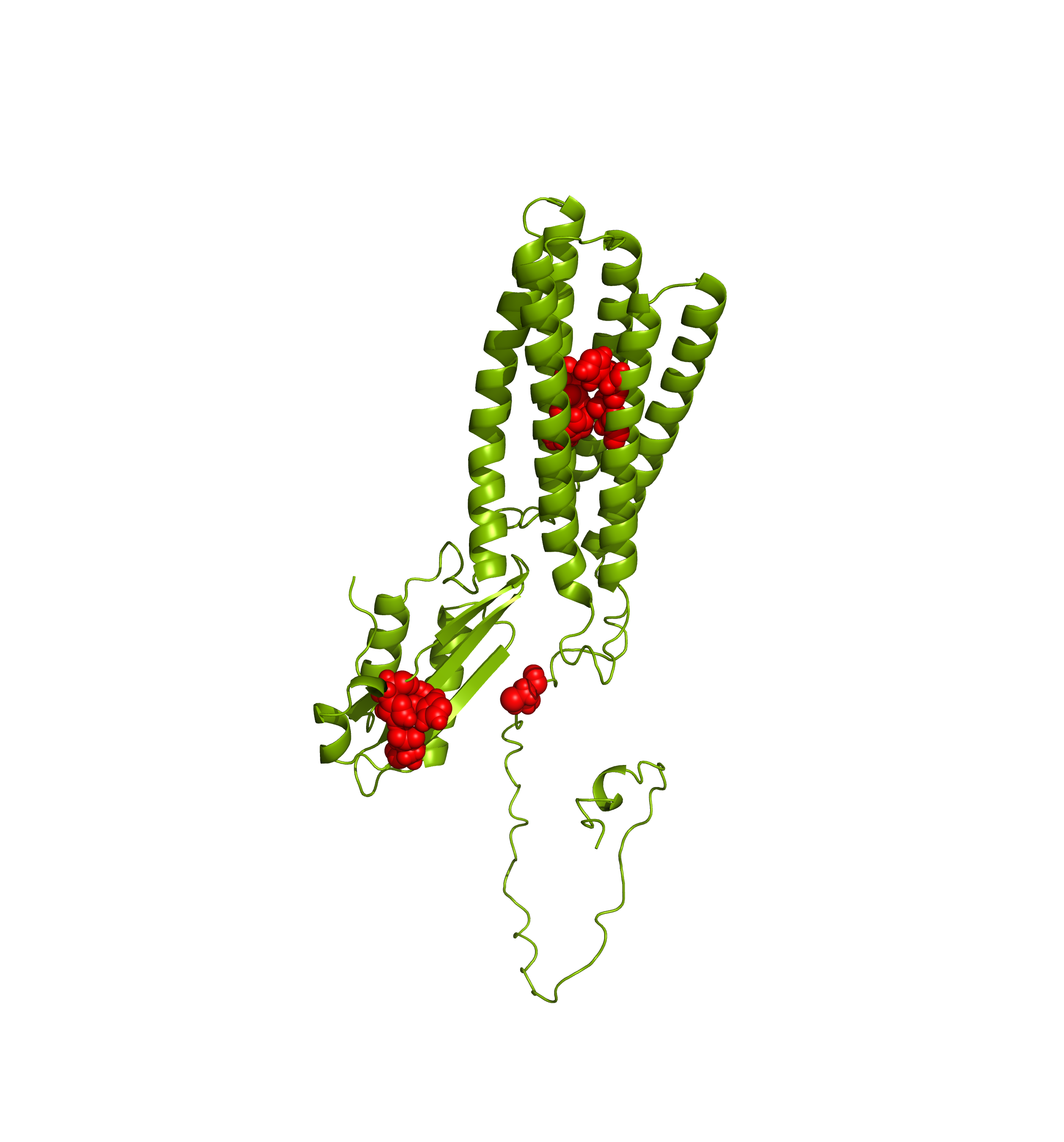
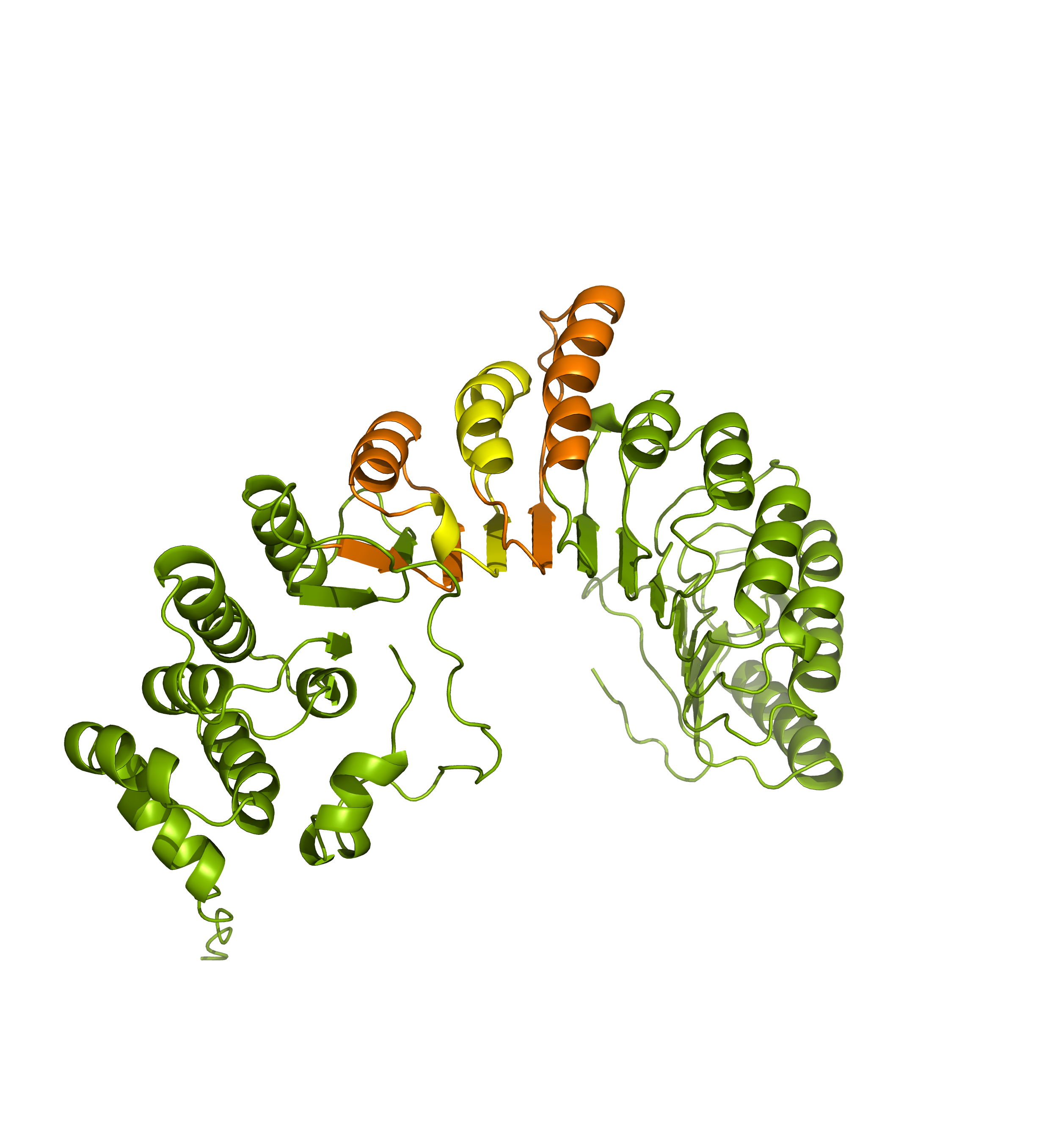
AlphaFoldDB model of PRIMEF26 (UniProt ID: H0Y7S4)
AlphaFoldDB currently (December 2024) stores a model of the UniProt entry H0Y7S4 that reflects version 2 of the H0Y7S4 sequence (changed in January 2024). Since then, an update to the UniProt entry led to a new version (version 3) of the sequence. Our alignment reveals that version 3 of H0Y7S4 sequence contains ~100 new residues 'inserted' at the beginning of the sequence that was modelled for AlphaFoldDB. In addition, the
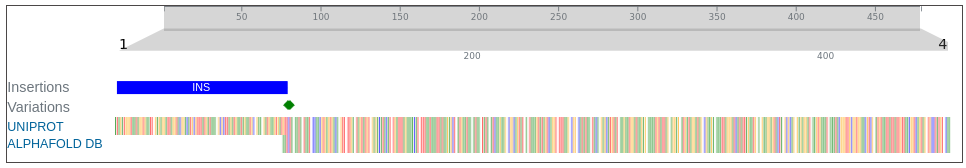
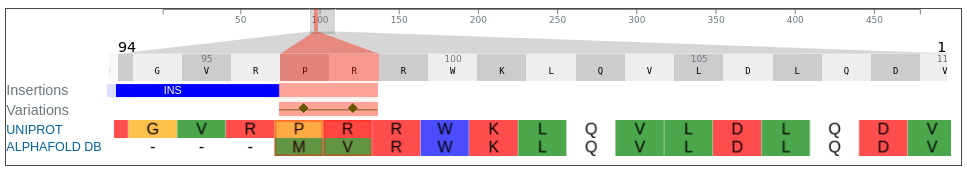
In addition, the first two residues (methionine and valine) modelled for AlphaFoldDB have since been updated in the UniProt to proline and arginine. They are annotated as variations on our dashoard and represented with green rectangles.
Comparing the structure deposited in AlphaFoldDB (purple, left) and the model of the up-to-date sequence (green, center) we see a somewhat preserved fold with the inserted sequence chunk not aligned with the AlphaFoldDB structure (structure alignment, on the right).
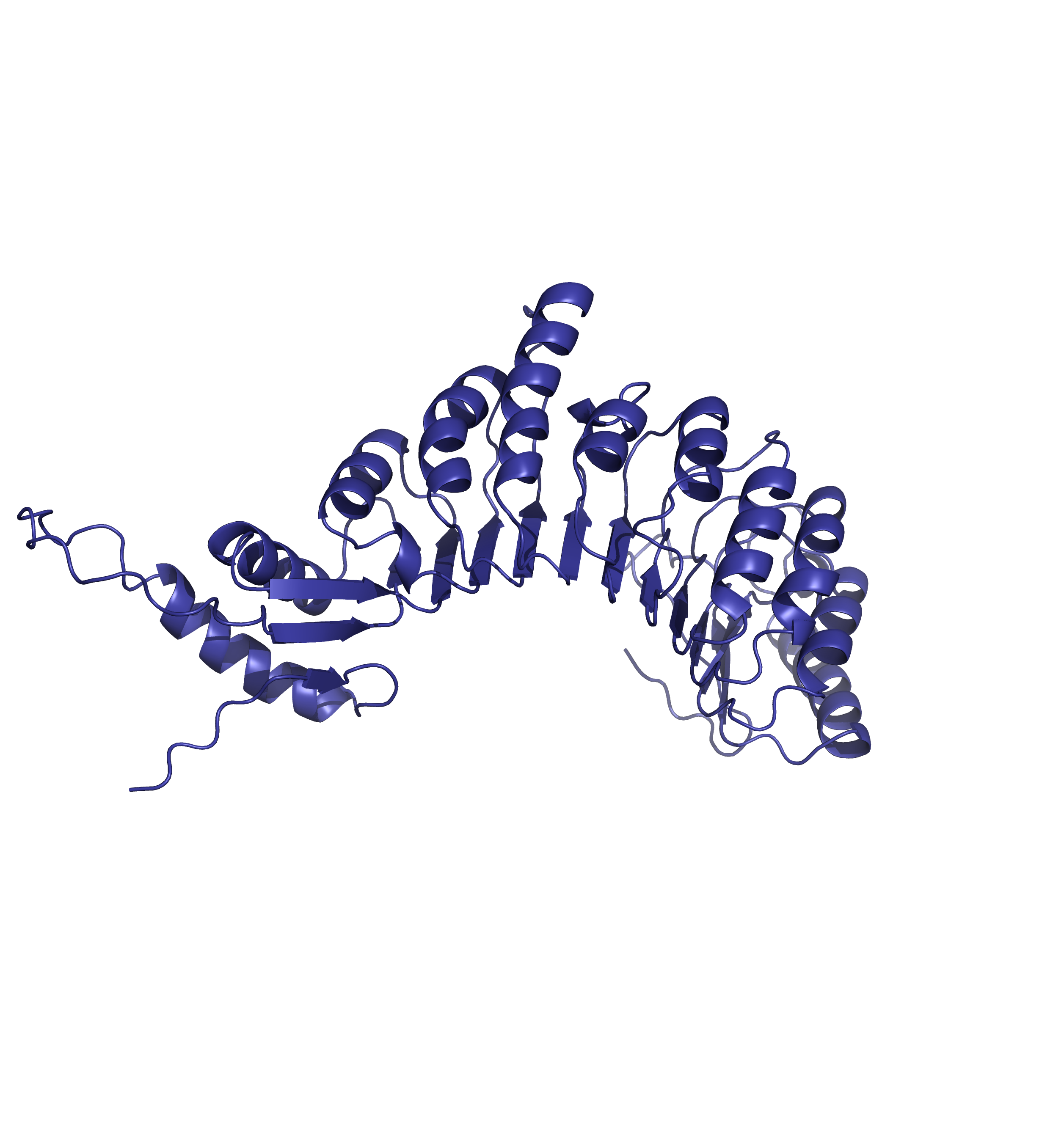
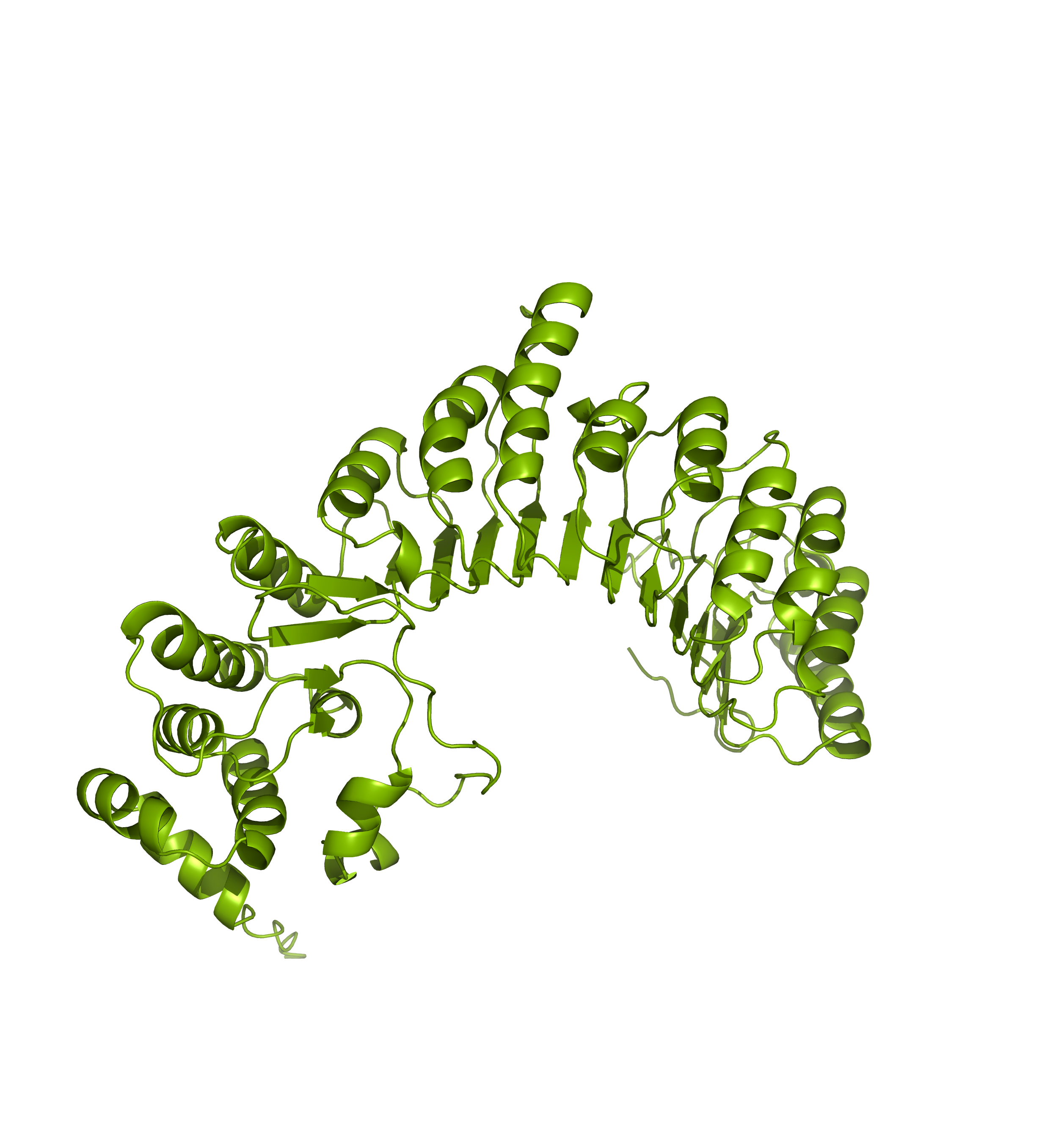
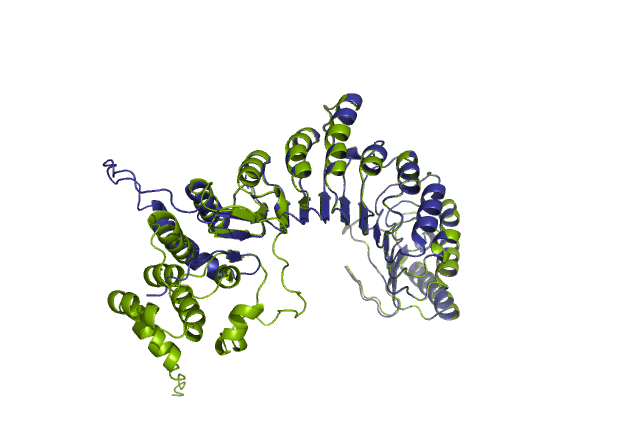
Forthermore, mapping annotations such domain annotations to the outdated models (highlighted below in orange) might result in misleading structural views as the domains in the AlphaFoldDB outdated structure (left) are shifted in relation to the actual domains in the updated model (right).
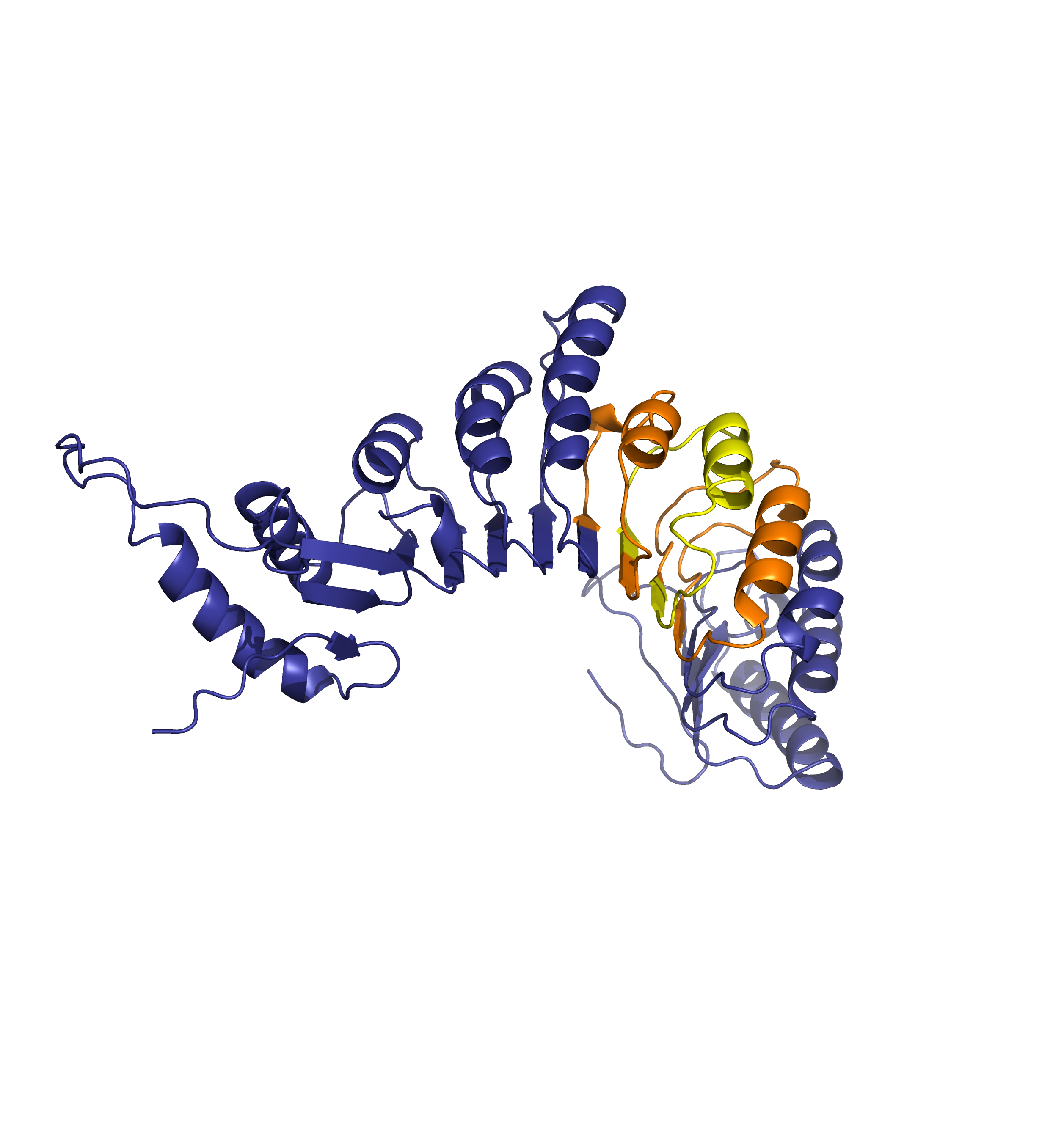
Time flies in bioinformatics - the aging of the AlphaFold DB. Manuscript in preparation