Documentation
Welcome to Missense3D, a tool for assessing the structural impact of amino acid substitutions in proteins. This guide will help you understand how to use the site and interpret your results.
What does Missense3D do?
Missense3D predicts the structural impact of missense variants on monomeric proteins and protein complexes, providing binary predictions as structurally ‘Damaging’ or ‘Neutral’ based on structural features. The tool includes two components: the standard Missense3D and Missense3D-PPI, which specifically evaluates variants located at protein–protein interaction interfaces.
Missense3D supports the analysis of both experimentally determined and computationally modelled structures, and is freely available to the scientific community. Variants can be submitted using their location in the UniProt protein sequence, their position in a 3D structure, or their genetic coordinates. The underlying methodology is described in detail here.What's on this page?
How to use Missense3D
How to input the variant coordinates and 3D structure coordinates
1. On the Missense3D front page, you can enter a variant using its location in the UniProt protein sequence, its position on a 3D structure, or its genetic coordinates.
Genetic Coordinates Input
Please select the tab marked "Genetic Variant Input" to input a variant using its genetic coordinates. Missense3D uses the Ensembl Variant Effect Predictor (VEP) to map human (GRCh38) genetic coordinates of missense variants to their corresponding protein coordinates.
Missense3D supports the following input formats:
- Ensembl default – chromosome, position, reference and alternate allele; e.g.,
5 43702652 43702652 T G. - Variant identifiers – dbSNP or other reference IDs; e.g.,
rs121913529. - HGVS notations – the format is according to the Human Genome Variation Society standards, e.g.,
ENST00000230882.9:c.344A>C. HGVS protein notations may also be used, provided that they unambiguously map to a single genomic change.
Missense3D analyses only protein-coding variants that map to the gene’s canonical transcript. After entering the genetic coordinates and pressing “Next” the tool returns detailed information for each variant, including the affected residue, predicted impact, and additional annotations.
Example Output:
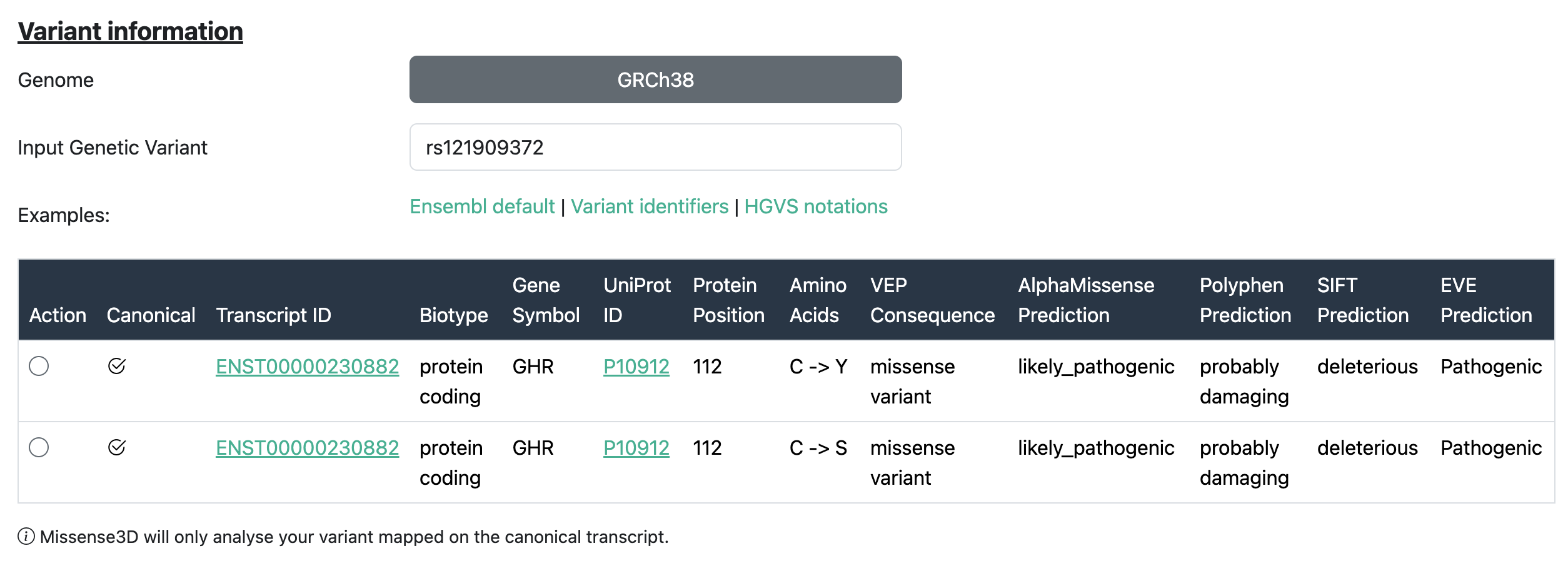
For each transcript considered, the following details are reported:
| Field | Description |
|---|---|
| Transcript ID | The Ensembl identifier for the transcript in which the variant is located (e.g., ENST00000380152). |
| Biotype | The classification of the transcript, such as protein-coding, pseudogene, etc. |
| Gene Symbol | The official gene symbol (e.g., TP53, BRCA1) associated with the transcript. |
| UniProt ID | The UniProt accession for the protein product of the transcript. |
| Protein Position | The position of the amino acid in the protein where the variant occurs. |
| Amino Acids | The reference and alternate amino acids affected by the variant (e.g., G/A). |
| VEP Consequence | The predicted effect of the variant on the transcript, such as missense variant, synonymous variant, etc. |
| AlphaMissense Prediction | Prediction from AlphaMissense indicating the potential impact of the variant on protein function. |
| PolyPhen Prediction | Functional impact prediction from PolyPhen-2: benign, possibly damaging, or probably damaging. |
| SIFT Prediction | SIFT score indicating whether the amino acid substitution affects protein function (e.g., tolerated or deleterious). |
| EVE Prediction | Prediction from the EVE model (Evolutionary model of Variant Effect) indicating variant pathogenicity. |
To learn more about VEP and variant annotation, visit the Ensembl documentation.
Subsequently, select one of the rows in the table and press Next. This will automatically populate all relevant fields in the "Protein Variant Input" tab.
Protein Variant Input
Select the tab marked "Protein Variant Input" to input a variant as a position within a UniProt sequence. The input requires the protein UniProt ID, the wild-type amino acid, its position and the amino acid substitution.
Then, select the 3D coordinates for the analysis of your variant. Missense3D supports the following input types:
- Choose a PDB ID and Chain ID – the user has to supply the PDB identifier and chain of an experimentally determined structure available from the Protein Data Bank.
- Use a tertiary structure AlphaFold model (Human only) – when this option is selected, we automatically retrieve the AlphaFold model corresponding to the query UniProt Id of a human protein. Currently, Missense3D only supports the automated retrieval of AlphaFold models of the human proteome. For the analysis of non-human variants using AlphaFold models, please use the “3D structure input“ option described below.
- Search for PPI complexes in the GWYRE Database (Human only) – this option allows for identifying 3D models of human protein complexes deposited in the GWYRE database.
Structure Variant Input
Please select the tab marked "3D Structure Input" to input a variant as a residue position on a 3D structure file. This input page requires knowledge of the position of the residue of interest in the 3D coordinate file (NOTE - residue numbering in 3D structures does not always match the residue numbering of the primary amino acid sequence). You can upload your own experimental or model structure (including AlphaFold models).
2. Structure Health Check
Before submitting your variant for analysis, a Structure Health Check is automatically performed. This step ensures that the residue of interest is well-resolved and suitable for structural interpretation. If the structure does not meet the quality criteria, the analysis may be skipped or limited. A full description of the evaluation criteria can be found here.
3. When you submit your request for variant analysis a waiting page will be displayed until the analysis completes.
The Waiting Page is displayed when your request has been submitted and the variant is being analysed. The page shows the time that your processing job was started. Please bear in mind that larger structures will take longer to analyse. When analysis is complete the display will automatically show the summary results page which will display a high-level summary of your results and a link to the detailed Results page. No user input required for this page.
Results Page
4. Once processing of your request has completed, you will be automatically directed to the summary Results page which shows any pathogenic structural features identified and includes a link to the detailed Results page for the current query.
The summary Results page is displayed once analysis of the variant structure is completed and an overall prediction is available.
- You can choose to download a copy of your results by selecting the "Download" button on the left right-side.
- The summary results line provides an overview of the analysis results which includes the Uniprot ID (if available) and the name of the structure file processed. These link to the corresponding Uniprot page and PDBe pages respectively (where appropriate).
- On the same line is confirmation of the variant position and the wild type and variant residue, followed by a summary of the structurally damaging features identified likely pathogenic damage to the structure, each damaging criterion is shown in white text on a red background.
- Finally, at the end of the summary line is a button marked "View Details" which will open the detailed Results page in a new tab.
5. The detailed Results page shows the results for the input variant at a given position in a UniProt sequence or at a given residue within a structure file. This page has a number of areas which contain a lot of information about the structure and the Missense3D analysis.
The Detailed Results page reports the detailed Missense3D analysis and allows the visualisation of the variant on the 3D structure. It opens in a new tab in your browser.
- You can choose to download a copy of your results by selecting the "Download" button near the top right of the page.
- Below this, on the left side of the screen, a JSmol window allows the display of an interactive model of the 3D protein structure. The structure can be rotated and zoomed interactively by using the mouse.
- To the right of this area is a summary of the variant information and a panel which offers coarse-grained control of the display. Advanced users can click on the 'console' link at the bottom of this panel to open up a console window for fine-grained control of the structure display.
- Below this is an area titled "Mutation Analysis Details" that contains the basic information of the variant and the Structural and Physicochemical Effects that Missense3D checked.
- Finally, near the bottom of the page is a section marked "Criteria Tested". If any criteria have indicated a pathogenic prediction, then they will be listed in information boxes at the top of this area marked as "Triggered Criteria".
- Criteria which did not indicate pathogenic effect are listed in information boxes below the others named "Checked Criteria" or "Not Applicable Criteria". The information boxes are expandable and contain more detailed information about the analysis results for that particular criterion.
Structural features analysed for each variant
The following structural criteria are used by Missense3D to determine whether a missense variant is predicted to be structurally damaging. Each feature captures a distinct structural disruption or alteration caused by the amino acid substitution:
| Structural Feature | Description |
|---|---|
| 1. Features considered damaging, irrespective of variant residue localisation | |
| Clash | A variant is flagged as having a clash if (1) the mutant MolProbity score is ≥
30.0, or (2) the difference between the mutant and wild-type MolProbity scores
is ≥ 18.0. The MolProbity score is computed using all atoms within a 20 Å sphere
centred on the Cα atom of the variant position. [column: Clash]
|
| Disallowed φ/ψ Angles | A variant is flagged as having disallowed phi/psi angles if the wild-type
residue has phi/psi angles within the allowed/favoured Ramachandran region,
while the mutant residue falls within an outlier region. [column:
Disallowed_phipsi_angle] |
| Secondary Structure Altered | The DSSP secondary structure assignment (summary character) differs between
wild-type and mutant. [column: Secondary_structure_altered] |
| Cavity Altered | Absolute difference in total cavity volume between wild type and mutant ≥ 70 ų.
[column: Cavity_altered] |
| Buried / Exposed Switch | Buried-to-exposed (RSA ↑ ≥ 0.09) or exposed-to-buried (RSA ↓ ≥ 0.15) transition.
[column: Buried_exposed_switch] |
| Cis Pro Replaced | Wild-type residue is Proline in cis configuration (ω angle between –45° and
+45°) and is replaced. [column: Cis_pro_replaced] |
| Gly in a Bend Replaced | Glycine in a DSSP-defined bend ('S') is replaced. [column:
Gly_in_a_bend] |
| Disulphide Breakage | Wild-type forms ≥1 disulphide bond, but mutant forms none. Cutoff: S–S ≤ 3.3 Å.
[column: Disulphide_bond_breakage] |
| 2. Features considered damaging when the variant residue is localised in a specific protein region (buried or interface) | |
| Buried/Interface Pro Introduced | Wild type is buried/interface and mutant is Proline. [column:
Buried_pro_introduced] |
| Buried/Interface Gly Replaced | Wild-type Glycine at buried/interface location is replaced. [column:
Buried_pro_introduced] |
| Buried/Interface Hydrophilic Introduced | Buried/interface hydrophobic residue is replaced with hydrophilic. [column:
Buried_hydrophylic_introduced] |
| Buried/Interface Charge Introduced | Uncharged buried/interface residue is replaced with a charged residue. [column:
Buried_charge_introduced] |
| Buried/Interface Charge Switched | Charged residue is replaced with opposite charge at buried/interface. [column:
Buried_charge_switched] |
| Buried/Interface Charge Replaced | Charged residue replaced by uncharged residue at buried/interface. [column:
Buried_charge_replaced] |
| Buried/Interface H-bond Breakage | Wild-type buried/interface residue forms H-bonds; mutant forms none. H-bond:
donor–acceptor ≤ 3.9 Å; H–acceptor ≤ 3.5 Å. [column:
Buried_Hbond_breakage] |
| Buried/Interface Salt Bridge Breakage | Wild-type buried/interface residue forms ≥1 salt bridge (≤ 5 Å); mutant forms
fewer. [column: Buried_salt_bridge_breakage] |
| 3. PPI features considered damaging when the variant residue is localised on an interface of a PPI complex | |
| Interface Tyr Replaced | Wild-type Tyrosine at interface is replaced with another residue. [column:
Interface_tyr_replaced] |
| Interface Trp Replaced | Wild-type Tryptophan at interface is replaced. [column:
Interface_tyr_replaced] |
| Interface H-bond Formation | Mutant forms ≥1 new intermolecular H-bond at interface not present in wild type.
H-bond: donor–acceptor ≤ 3.9 Å; H–acceptor ≤ 3.5 Å. [column:
Interface_Hbond_formation] |
| Interface Salt Bridge Formation | Mutant forms more intermolecular salt bridges at interface than wild type
(cutoff ≤ 5 Å). [column: Interface_salt_bridge_formation] |
Additionally:
Structure Health Check
Our structure health check primarily relies on 3DSeqCheck, a web-based tool that verifies sequence consistency between a 3D structure file and its corresponding UniProt entry. This ensures that users do not receive incorrect results due to mapping errors or low-quality structural data. You can learn more about how it works here.
The criteria tested by the Structure Health Check are the following:
| Sequence-Based Criteria | |
|---|---|
| Feature | Description |
| UniProt ID | The unique accession identifier for the protein in the UniProt database. |
| Residue Number | The specific position in the protein sequence being evaluated. |
| Wild Type Amino Acid | The expected amino acid at the given position according to UniProt. |
| Residue match with UniProt | Check if the amino acid at the specified position matches the UniProt reference sequence. |
| Structure-Based Criteria | |
|---|---|
| Feature | Description |
| PDB ID | The Protein Data Bank (PDB) identifier for the 3D structure. |
| Chain | The specific chain in the PDB structure that is analysed. |
| Method | The experimental technique used to determine the 3D structure (e.g., X-ray crystallography). Structures derived from NMR or Cryo-EM are flagged due to potential limitations in resolution or completeness. |
| Resolution | The resolution of the structure in Ångströms; lower values indicate higher quality. Structures with a resolution above 3 Å are flagged and not recommended for use. |
| Residue exists in the structure | Checks whether the specified residue position exists in the structure data. |
| Residue match in structure | Checks whether the UniProt residue can be mapped to the structure correctly. |
| UniProt Sequence Length | Total number of amino acids in the UniProt reference sequence. |
| PDB Sequence Length | Total number of residues present in the structural model. |
| Coverage | Percentage of the UniProt sequence that is mapped in the structure. |
| AlphaFold Criteria | |
|---|---|
| Feature | Description |
| AlphaFold model available | Indicates whether an AlphaFold model exists (only for human proteins under 2,700 amino acids). |
| pLDDT | AlphaFold's per-residue confidence score (0–100), where higher values indicate greater confidence. Scores below 70 are flagged as low-confidence regions. |
The JSmol console
1. How to visualize multiple residues using the JSmol console
On the JSmol structure viewer, right-click to open a menu and select Console or alternatively click on the JSmol Console button; the JSmol console will then appear, displaying a command interface.
2. Examples of commands that can be used from the JSmol console for model structures
To interact with the structure using the JSmol console, use the following commands:
select 60,70; colour darkgrey; wireframe 1.5— selects residues 60 and 70, colors them dark grey, and displays them in wireframe.colour background light grey— changes the background color to light grey.select *— selects the entire structure.wireframe 0.5— shows the sidechains of all residues with default thickness (use other values like 1.5 or 0.2 to change thickness).wireframe off— hides all sidechains.
Press Enter after typing each command in the console.
3. Examples of commands that can be used from the JSmol console for PDB structures
IMPORTANT: The numbering of amino acids in PDB structures is often arbitrary and may not match UniProt numbering. When using the console to select residues, it's essential to know the UniProt-to-PDB residue mapping and the chain ID of the protein of interest.
Use the following commands in the JSmol console:
select :a— selects chain A only.colour light green— colors the selected chain light green.select :c and 40— selects residue 40 in chain C.colour darkgrey; wireframe 0.5— colors the selected residue dark grey and displays its side chain.select :c and 40,50; colour darkgrey; wireframe 0.5— selects residues 40 and 50 in chain C, colors them dark grey, and displays their side chains.select :c and pro— selects all proline residues in chain C.wireframe 0.5— displays the side chains of the selected residues.colour darkgrey— colors the selected residues dark grey.select hydrophobic— selects all hydrophobic residues.- To add residue labels, right-click on the structure viewer, select Color → Labels, and choose a label color (e.g. White).
More advanced command-line options can be found online. However, please note that Missense3D is not intended for full molecular visualization. For advanced visual tasks, we recommend using dedicated molecular viewers such as PyMOL or EzMol.